4. Example scripts¶
This section describes various example scripts to demonstrate pyGCluster
4.1. Functionality check¶
Note
After installation, please run the script test_pyGCluster.py from the exampleScripts folder to check if pyGCluster was installed properly.
Testscript to demonstrate functionality of pyGCluster
A synthetic dataset is used to check the correct installation of pyGCluster. This dataset cotains 10 ratios (Gene 0-9) which were randonmly sampled between 39.5 and 40.5 in 0.1 steps with a low standard deviation (randonmly sampled between 0.1 and 1) and 90 ratios (Gene 10-99) which were randonmly sampled between 3 and 7 in 0.1 steps with a high standard deviation (randonmly sampled between 0.1 and 5)
5000 iterations are performed and the presence of the most frequent cluster is checked.
This cluster should contain the Genes 0 to 9.
Usage:
./test_pyGCluster.py
When the iteration has finished (this should normally take not longer than 20 seconds), the script asks if you want to stop the iteration process or continue:
iter_max reached. See convergence plot. Stopping re-sampling if not defined
otherwise ...
... plot of convergence finished.
See plot in "../exampleFiles/functionalityCheck/convergence_plot.pdf".
Enter how many iterations you would like to continue.
(Has to be a multiple of iterstep = 5000)
(enter "0" to stop resampling.)
(enter "-1" to resample until iter_max (= 5000) is reached.)
Enter a number ...
Please enter 0 and hit enter (The script will stop and the test will finish).
The results are saved into the folder functionalityCheck.
Additionally expression maps and expression profiles are plotted.
4.2. Testscripts to demonstrate pyGCluster¶
The basic script utilizes the function pyGCluster.Cluster.do_it_all() whereas the advanced script utilizes single steps to perform the clustering
4.2.1. Basic script¶
Testscript to demonstrate functionality of pyGCluster
This script imports the data of Hoehner et al. (2013) and executes pyGCluster with 250,000 iterations of resampling. pyGCluster will evoke 4 threads (if possible), which each require approx. 1.5GB RAM. Please make sure you have enough RAM available (4 threads in all require approx. 6GB RAM). Duration will be approx. 2 hours to complete 250,000 iterations on 4 threads.
Usage:
./basicClusterHoehnerExampleData.py <pathToExampleFile>
If this script is executed in folder pyGCluster/exampleScripts, the command would be:
./basicClusterHoehnerExampleData.py ../exampleFiles/hoehner_dataset.csv
The results are saved in ”.../pyGCluster/exampleScripts/hoehner_example_run/”.
4.2.2. Advanced script¶
Testscript to demonstrate functionality of pyGCluster
This script imports the data of Hoehner et al. (2013) and executes pyGCluster with 250,000 iterations of resampling. pyGCluster will evoke 4 threads (if possible), which each require approx. 1.5GB RAM. Please make sure you have enough RAM available (4 threads in all require approx. 6GB RAM). Duration will be approx. 2 hours to complete 250,000 iterations on 4 threads.
Usage:
./advancedClusterHoehnerExampleData.py <pathToExampleFile>
If this script is executed in folder pyGCluster/exampleScripts, the command would be:
./advancedClusterHoehnerExampleData.py ../exampleFiles/hoehner_dataset.csv
The results are saved in ”.../pyGCluster/exampleScripts/hoehner_example_run/”.
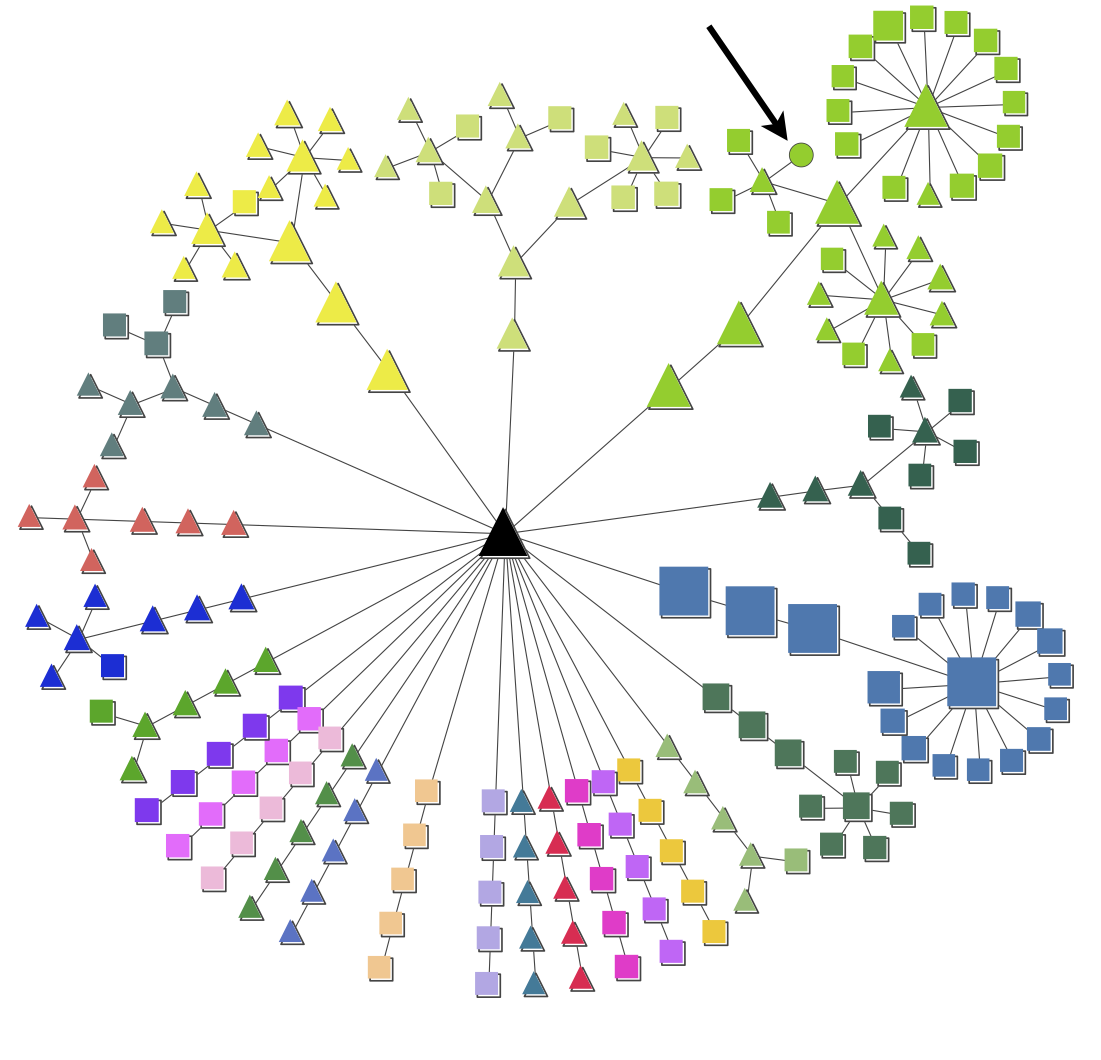
4.3. Plot communities from a pyGCluster pkl¶
Plot communities of a given pyGCluster pkl file. All output will be directed into the folder, wehre the input data is located
The top_X_clusters option can be used to use the top X clusters for community determination. The threshold_4_the_lowest_max_freq option define the threshold for the maximum frequency of the clusters which should be incoporated into the community determination.
Default values are:
-threshold_4_the_lowest_max_freq=0.005
Usage:
./plotCommunities.py <pathTopyGClusterPickle>
optional:
./plotCommunities.py <pathTopyGClusterPickle> <threshold_4_the_lowest_max_freq=0.005>
OR <top_X_clusters=100>
4.4. Retrieve pyGCluster pkl info¶
Get some information from a pyGCluster pkl object
Usage:
./getpyGPickleInfo.py <MERGED_pyGCluster_pkl_object>
4.5. Plot simple expression map¶
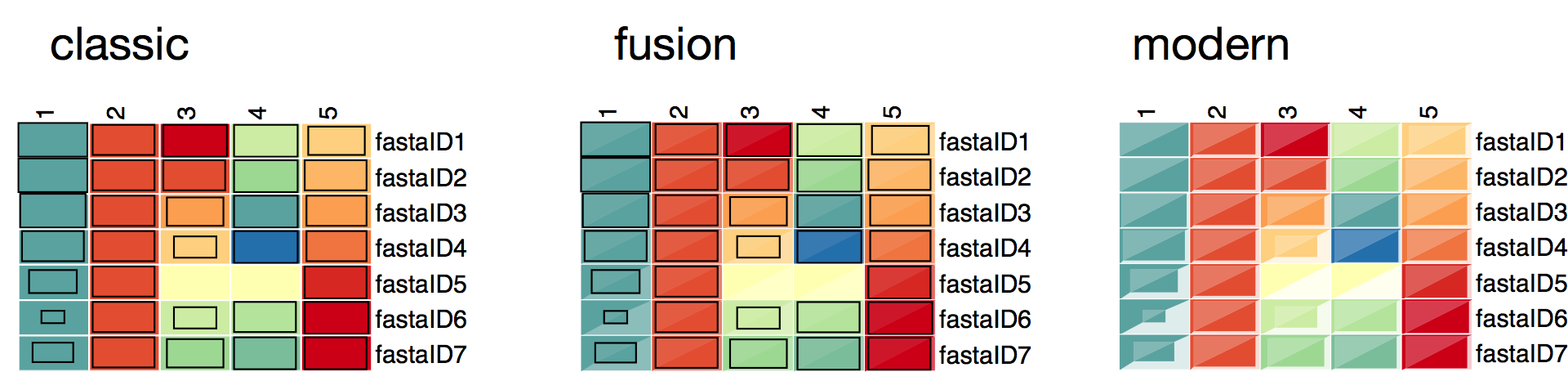
Testscript to plot a simple heatmap.
This can be used to visualize the different box styles for the expression maps and to test the plotting function.
This can be also used as a basis to visualize own datasets by simply defining the ‘data’ dictionary in this script.
Usage:
./simpleExpressionMap.py